RNA-Seq and SHAPE chemistry
A collaboration with the Granneman lab is developing probabilistic techniques for analysing next-generation RNA-Seq data. SHAPE (selective 2' hydroxyl acylation analysed by primer extension) chemistry is based on the discovery that, in RNA, the 2' hydroxyl is unreactive at nucleotides constrained by base pairing, but reactive at flexible positions. After the addition of a chemical reagent that binds selectively to the RNA, reverse transcription generates cDNA which is then analysed by NGS yielding information about RNA structure. The Granneman lab is investigating the RNA folding steps that take place in ribosomal RNA during ribosome assembly in yeast.
Regulatory sequences
The MEME algorithm has been successfully applied to identify regulatory sequences in magnetic bacteria by Alastair Kilpatrick (thesis), in collaboration with Bruce Ward. Known regulators have been recovered from upstream regions of the genomes of several bacteria, and new potential motifs have been predicted. Extensions to the EM algorithm underlying MEME have been evaluated, paving the way for future work.
Data mining
Luna De Ferrari has recently published the EnzML technique for assigning enzymatic function to the proteins using their InterPro signatures as a feature space. EnzML can re-annotate entire proteomes with subset accuracy ranging from 87% for A. thaliana and reaching 97% for E. coli.
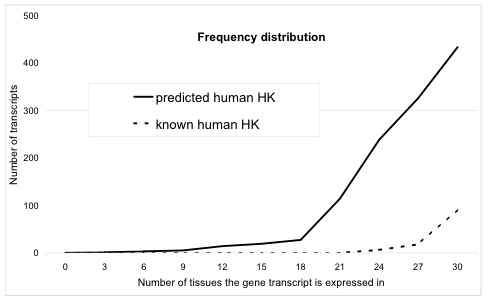
A Bayesian approach capable of aggregating genomic data to predict whether a gene has a housekeeping function or not has shown great promise. Luna De Ferrari has been able to predict over 550 human and over 2000 mouse genes with a high probability of having a housekeeping role (thesis), see figure centre right.
An evolutionary approach to classifying microarray data according to the original cell lines of the samples, e.g. the AML and ALL subtypes of leukaemia, is shown by Thanyaluk Jirapech-Umpai to yield an accuracy of 98% (thesis). Genes inferred by the algorithm to be highly discriminatory include those known to indicate leukaemia.
Gene network learning
The performance of an optimal gene network inference algorithm has been
evaluated by Shivani Puri
(thesis).
Using simulated data, performance over a range of sample
sizes and noise values was analysed.
Alternative gene network scoring functions,
and the use of wavelet transforms to analyse temporal data have
been investigated by Sock Leh (June) Tee
(
thesis).
Initial results using classifier learning as a feature selection
step in the process of analysing microarray data have shown promise.
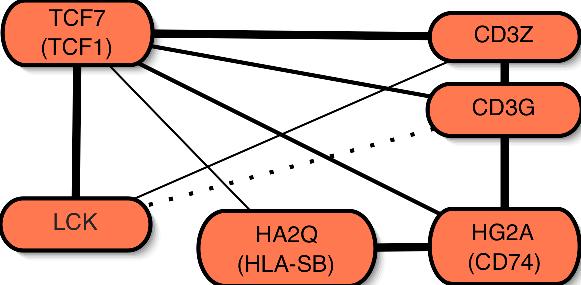
The inferred T-cell network (bottom right) was derived from leukaemia data
by combining optimal networks using a bootstrapping approach to resample from
from the limited data available.