Dizzy Beats:
Parameter inference and model comparison in computational systems biology
DizzyBeats-beta available for
download
i) Java jar application (all platforms)
ii) Mac OS X application
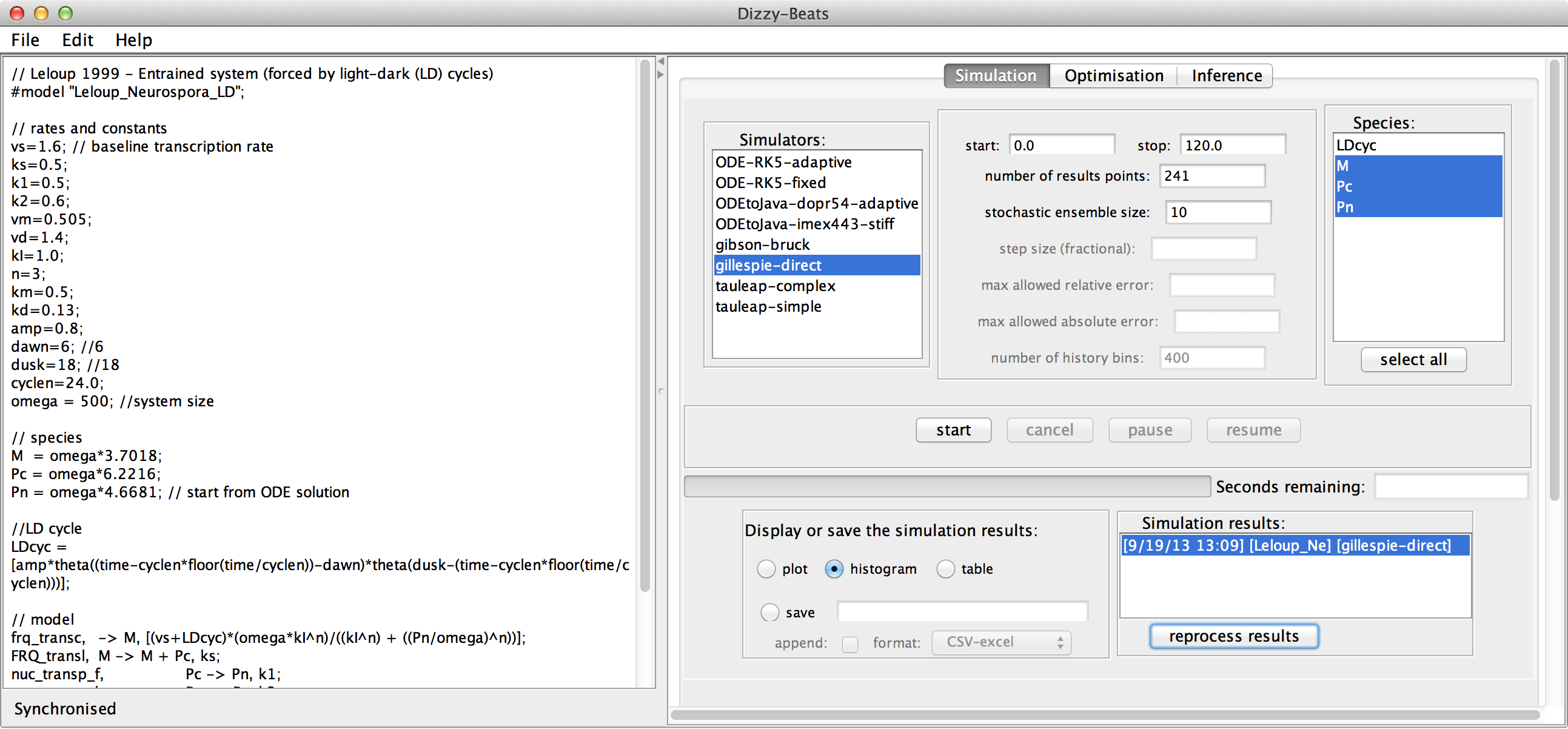
The interface of Dizzy has been revised to show the model
in the left panel and the simulator controls in the right
panel. This allows models to be edited without moving
between windows.
A new histogram display has been added.
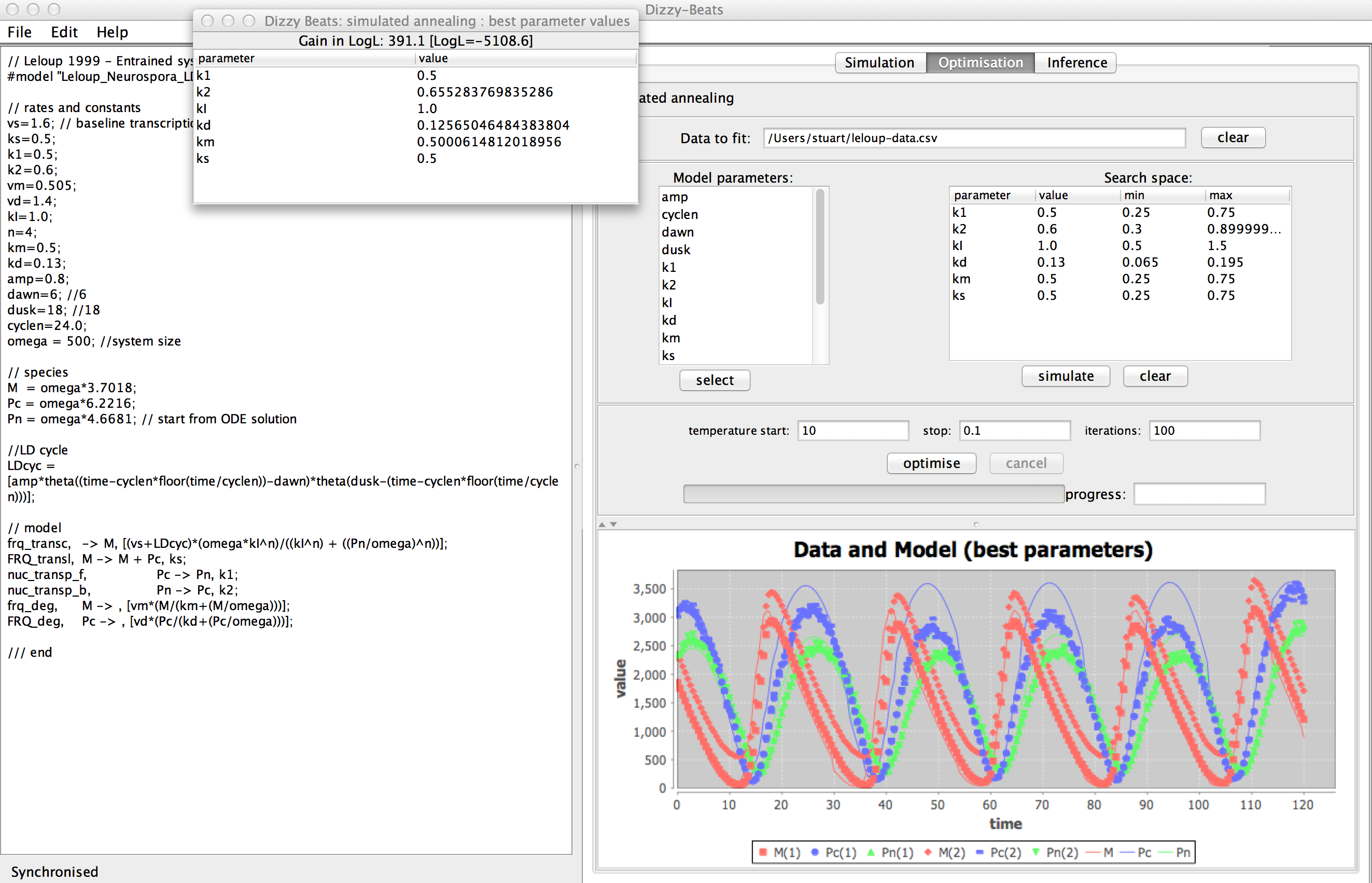
A simulated annealing algorithm allows selected model
parameters to be fitted to data.
A generic likelihood function based on the L1 norm is used to assess fit, this
requires only that there be two or more replicates in the
data for each data series.
The Simulation panel allows data to be loaded, data must be formatted in the commonly-used CSV format.
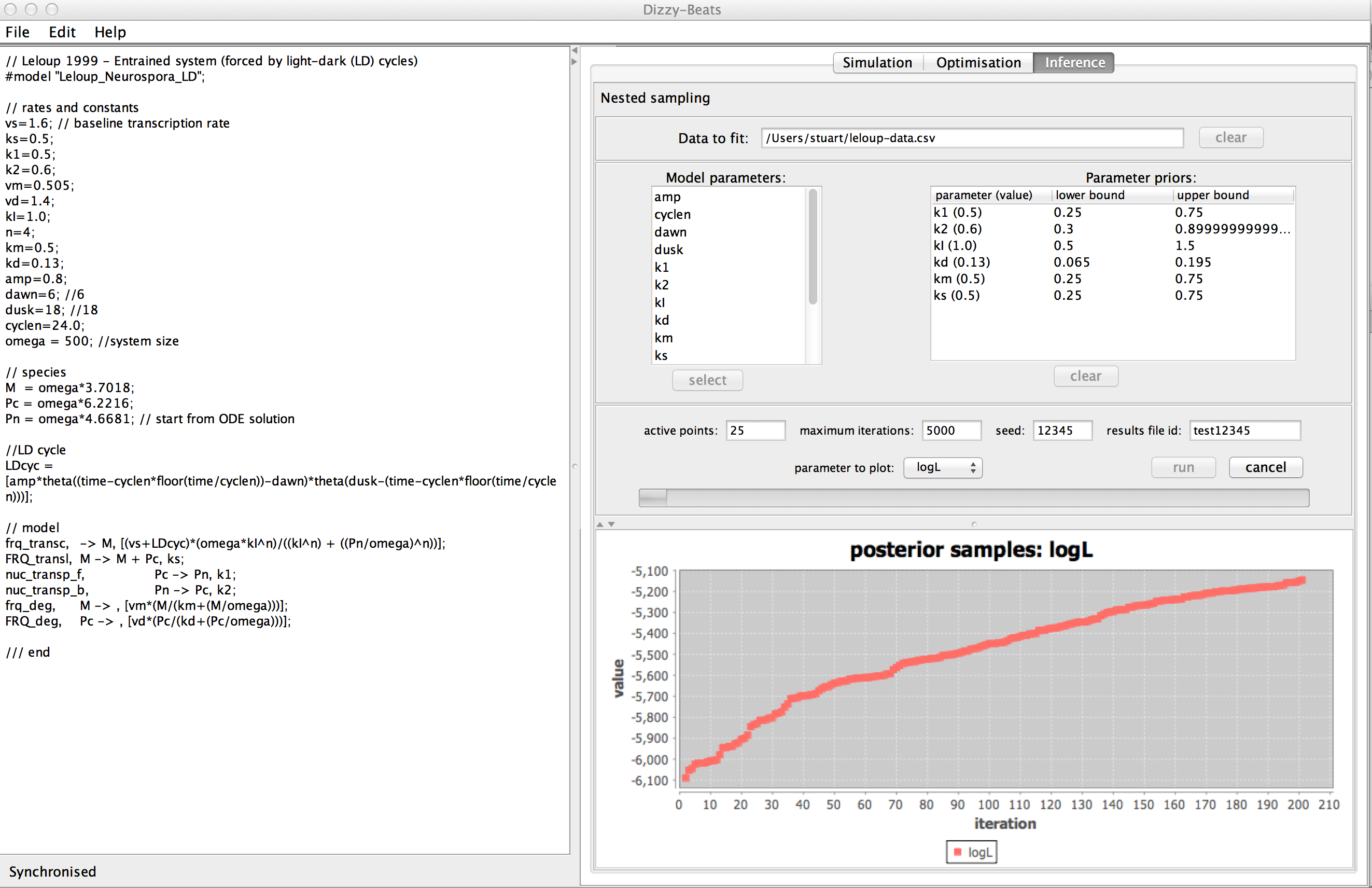
Nested sampling performs the Bayesian Evidence calculation
for model comparison, and generates estimates of the parameter
means and standard deviations. As with the optimiser, the
likelihood function requires only that there be two or
more replicates in the data. The Inference panel allows
the data to be loaded and inspected, and displays
diagnostic information as nested sampling runs. Users can
monitor the gain in log likelihood, the gain (then loss)
in the log width associated with each posterior sample,
and can view the pattern of posterior parameter
values. These plots are updated as the algorithm runs, and
the user can inspect any element of the posterior data
using the GUI.